- Introduction
- Pre-processing raster predictors
- Preparation of data for model projections
- Project selected models to multiple scenarios
- Import rasters resulting from projections
Introduction
Once selected models have been fit using fit_selected(),
projections to single or multiple scenarios can be performed. The
project_selected() function is designed for projections to
multiple scenarios.
To project using the selected models, a fitted_models
object is required. For detailed information on model fitting, please
consult the vignette Fit and Explore
Selected Models. The fitted_models object generated in
that vignette is included as an example dataset within the package.
Let’s load it.
#Load packages
library(kuenm2)
library(terra)
#> terra 1.8.60
#Import calib_results_maxnet
data("fitted_model_maxnet", package = "kuenm2")
#Print calibration result
fitted_model_maxnet
#> fitted_models object summary
#> ============================
#> Species: Myrcia hatschbachii
#> Algortihm: maxnet
#> Number of fitted models: 2
#> Models fitted with 4 replicatesPre-processing raster predictors
While predicting models for a single scenario requires a single
SpatRaster object containing the predictor variables (as
detailed in Predict models to single
scenario), projecting models to multiple scenarios necessitates a
folder that stores the predictor variables for each scenario.
These folders must be organized in a specific hierarchical manner: a root directory should contain nested folders representing different scenarios, with the raster variables stored within. At the first level inside the root folder, subfolders should correspond to distinct time periods (e.g., future years like “2070” or “2100,” or past periods such as “Mid-holocene” or “LGM”). Within each period folder, if applicable, you should include subfolders for each emission scenario (e.g., “ssp126”, “ssp585”). Finally, within each emission scenario or time period folder, include a separate folder for each General Circulation Model (GCM) (e.g., “BCC-CSM2-MR”, “MIROC6”). This structured organization enables the function to automatically access and process the data according to period, emission scenario, and GCM.
Organize and structure future climate variables from WorldClim
The package provides a function to import future climate variables downloaded from WorldClim (version 2.1). This function renames the files and organizes them into folders categorized by period/year, emission scenario (Shared Socioeconomic Pathways; SSPs), and General Circulation Model (GCM). This simplifies the preparation of climate data, ensuring all required variables are properly structured for modeling projections.
To use this function, download the future raster variables from WorldClim 2.1 and save them all within the same folder. DO NOT rename the files or the variables, as the function relies on the patterns provided in the original files to work properly.
The package also provides an example of raw variables downloaded from WorldClim 2.1. This example includes bioclimatic predictions for the periods “2041-2060” and “2081-2100”, for two SSPs (125 and 585) and two GCMs (ACCESS-CM2 and MIROC6), at 10 arc-minutes resolution.
# See raster files with future predictors provided as example
# The data is located in the "inst/extdata" folder.
in_dir <- system.file("extdata", package = "kuenm2")
list.files(in_dir)
#> [1] "bias_file.tif"
#> [2] "CHELSA_LGM_CCSM4.tif"
#> [3] "CHELSA_LGM_CNRM-CM5.tif"
#> [4] "CHELSA_LGM_FGOALS-g2.tif"
#> [5] "CHELSA_LGM_IPSL-CM5A-LR.tif"
#> [6] "CHELSA_LGM_MIROC-ESM.tif"
#> [7] "CHELSA_LGM_MPI-ESM-P.tif"
#> [8] "CHELSA_LGM_MRI-CGCM3.tif"
#> [9] "Current_CHELSA.tif"
#> [10] "Current_variables.tif"
#> [11] "m.gpkg"
#> [12] "wc2.1_10m_bioc_ACCESS-CM2_ssp126_2041-2060.tif"
#> [13] "wc2.1_10m_bioc_ACCESS-CM2_ssp126_2081-2100.tif"
#> [14] "wc2.1_10m_bioc_ACCESS-CM2_ssp585_2041-2060.tif"
#> [15] "wc2.1_10m_bioc_ACCESS-CM2_ssp585_2081-2100.tif"
#> [16] "wc2.1_10m_bioc_MIROC6_ssp126_2041-2060.tif"
#> [17] "wc2.1_10m_bioc_MIROC6_ssp126_2081-2100.tif"
#> [18] "wc2.1_10m_bioc_MIROC6_ssp585_2041-2060.tif"
#> [19] "wc2.1_10m_bioc_MIROC6_ssp585_2081-2100.tif"Note that all variables are in the same folder and retain the
original names provided by WorldClim. You can download these variables
directly from WorldClim
or by using the geodata R package:
#Install geodata if necessary
if(!require("geodata")){
install.packages("geodata")
}
#Load geodata
library(geodata)
#Create folder to save the raster files
#Here, in a temporary directory
geodata_dir <- file.path(tempdir(), "Future_worldclim")
dir.create(geodata_dir)
#Define GCMs, SSPs and time periods
gcms <- c("ACCESS-CM2", "MIROC6")
ssps <- c("126", "585")
periods <- c("2041-2060", "2061-2080")
#Create a grid of combination of periods, ssps and gcms
g <- expand.grid("period" = periods, "ssps" = ssps, "gcms" = gcms)
g #Each line is a specific scenario for future
#Loop to download variables for each scenario
lapply(1:nrow(g), function(i){
cmip6_world(model = g$gcms[i],
ssp = g$ssps[i],
time = g$period[i],
var = "bioc",
res = 10, path = geodata_dir)
}) #It will take a while...Let’s check the variables inside the “geodata_dir” folder:
list.files(geodata_dir, recursive = TRUE)
#> [1] "climate/wc2.1_10m/wc2.1_10m_bioc_ACCESS-CM2_ssp126_2041-2060.tif"
#> [2] "climate/wc2.1_10m/wc2.1_10m_bioc_ACCESS-CM2_ssp126_2061-2080.tif"
#> [3] "climate/wc2.1_10m/wc2.1_10m_bioc_ACCESS-CM2_ssp585_2041-2060.tif"
#> [4] "climate/wc2.1_10m/wc2.1_10m_bioc_ACCESS-CM2_ssp585_2061-2080.tif"
#> [5] "climate/wc2.1_10m/wc2.1_10m_bioc_MIROC6_ssp126_2041-2060.tif"
#> [6] "climate/wc2.1_10m/wc2.1_10m_bioc_MIROC6_ssp126_2061-2080.tif"
#> [7] "climate/wc2.1_10m/wc2.1_10m_bioc_MIROC6_ssp585_2041-2060.tif"
#> [8] "climate/wc2.1_10m/wc2.1_10m_bioc_MIROC6_ssp585_2061-2080.tif"
#>
#> #Set climate as input directory
#> in_dir <- file.path(geodata_dir, "climate")Now, we can organize and structure the files using the
organize_future_worldclim function.
Format for renaming
An important argument is name_format, which defines the
format for renaming variables. The names of the variables in the
SpatRaster must precisely match those used for model
calibration or when running PCA (if do_pca = TRUE was set
in the prepare_data() function; see Prepare Data for Model Calibration for more
details).
Therefore, if the variables used to calibrate the models are named “bio_1”, “bio_2”, etc., the variables in the future raster layers must also be named “bio_1”, “bio_2”, etc. However, if the variables have a different pattern, such as starting with uppercase letters and using zeros before single-digit numbers (e.g., “Bio_01”, “Bio_02”, etc.), they must be named “Bio_01”, “Bio_02”, etc. The function provides four options:
-
"bio_": Variables will be renamed tobio_1,bio_2,bio_3,bio_10, etc. -
"bio_0": Variables will be renamed tobio_01,bio_02,bio_03,bio_10, etc. -
"Bio_": Variables will be renamed toBio_1,Bio_2,Bio_3,Bio_10, etc. -
"Bio_0": Variables will be renamed toBio_01,Bio_02,Bio_03,Bio_10, etc.
Let’s check how the variables were named in our
fitted_model:
fitted_model_maxnet$continuous_variables
#> [1] "bio_1" "bio_7" "bio_12" "bio_15"The variables follows the standards of the first option
("bio_").
Fixed variables
When predicting for other times, you can assume that some variables
will be static (i.e., they remain unchanged in the projected scenarios).
The fixed_variables argument allows you to append static
variables alongside the bioclimatic variables.
Here, let’s assume soilType will remain static in the
future scenarios:
# Import raster layers (same used to calibrate and fit final models)
var <- rast(system.file("extdata", "Current_variables.tif", package = "kuenm2"))
#Get soilType
soiltype <- var$SoilTypeOrganize and structure WorldClim files
Now, let’s organize the WorldClim files with the
organize_future_worldclim() function:
#Create folder to save structured files
out_dir_future <- file.path(tempdir(), "Future_raw") #Here, in a temporary directory
#Organize
organize_future_worldclim(input_dir = in_dir, #Path to the raw variables from WorldClim
output_dir = out_dir_future,
name_format = "bio_", #Name format
fixed_variables = var$SoilType) #Static variables
#> | | | 0% | |========= | 12% | |================== | 25% | |========================== | 38% | |=================================== | 50% | |============================================ | 62% | |==================================================== | 75% | |============================================================= | 88% | |======================================================================| 100%
#>
#> Variables successfully organized in directory:
#> /tmp/Rtmpw1qyx4/Future_raw
# Check files organized
dir(out_dir_future, recursive = TRUE)
#> [1] "2041-2060/ssp126/ACCESS-CM2/Variables.tif"
#> [2] "2041-2060/ssp126/MIROC6/Variables.tif"
#> [3] "2041-2060/ssp585/ACCESS-CM2/Variables.tif"
#> [4] "2041-2060/ssp585/MIROC6/Variables.tif"
#> [5] "2081-2100/ssp126/ACCESS-CM2/Variables.tif"
#> [6] "2081-2100/ssp126/MIROC6/Variables.tif"
#> [7] "2081-2100/ssp585/ACCESS-CM2/Variables.tif"
#> [8] "2081-2100/ssp585/MIROC6/Variables.tif"We can check the files structured hierarchically in nested folders
using the dir_tree() function from the fs
package:
#Install package if necessary
if(!require("fs")){
install.packages("fs")
}
dir_tree(out_dir_future)
#> Temp\RtmpkhmGWN/Future_raw
#> ├── 2041-2060
#> │ ├── ssp126
#> │ │ ├── ACCESS-CM2
#> │ │ │ └── Variables.tif
#> │ │ └── MIROC6
#> │ │ └── Variables.tif
#> │ └── ssp585
#> │ ├── ACCESS-CM2
#> │ │ └── Variables.tif
#> │ └── MIROC6
#> │ └── Variables.tif
#> └── 2081-2100
#> ├── ssp126
#> │ ├── ACCESS-CM2
#> │ │ └── Variables.tif
#> │ └── MIROC6
#> │ └── Variables.tif
#> └── ssp585
#> ├── ACCESS-CM2
#> │ └── Variables.tif
#> └── MIROC6
#> └── Variables.tifAfter organizing variables, the next step is to create the
prepared_projection object.
Preparation of data for model projections
Now, let’s prepare data for model projections across multiple scenarios, storing the paths to the rasters representing each scenario.
In contrast to predict_selected(), which requires a
SpatRaster object, we need the paths to the folders where
the raster files are stored. This includes the variables for the present
time, which were used to calibrate and fit the models.
Currently, we only have the future climate files. The present-day predictor variables must reside in the same root directory as the processed future variables. Let’s copy the rasters used for model calibration and fitting to this folder:
# Create a "Current_raw" folder in a temporary directory
# and copy the rawvariables there.
out_dir_current <- file.path(tempdir(), "Current_raw")
dir.create(out_dir_current, recursive = TRUE)
# Save current variables in temporary directory
terra::writeRaster(var, file.path(out_dir_current, "Variables.tif"))
#Check folder
list.files(out_dir_current)
#> [1] "Variables.tif"Now, we can prepare the data for projections. In addition to storing
the paths to the variables for each scenario, the function also verifies
if all variables used to fit the final models are available across all
scenarios. To perform this check, you need to provide either the
fitted_models object you intend to use for projection or
simply the variable names. We strongly suggest using the
fitted_models object to minimize projection errors.
We also need to define the root directory containing the scenarios for projection (present, past, and/or future), along with additional information regarding time periods, SSPs, and GCMs.
# Prepare projections using fitted models to check variables
pr <- prepare_projection(models = fitted_model_maxnet,
present_dir = out_dir_current, #Directory with present-day variables
past_dir = NULL, #NULL because we won't project to the past
past_period = NULL, #NULL because we won't project to the past
past_gcm = NULL, #NULL because we won't project to the past
future_dir = out_dir_future, #Directory with future variables
future_period = c("2041-2060", "2081-2100"),
future_pscen = c("ssp126", "ssp585"),
future_gcm = c("ACCESS-CM2", "MIROC6"))When we print the projection_data object, it summarizes
all the scenarios we will predict and also shows the root directory
where the predictor rasters are stored:
pr
#> projection_data object summary
#> =============================
#> Variables prepared to project models for Present and Future
#> Future projections contain the following periods, scenarios and GCMs:
#> - Periods: 2041-2060 | 2081-2100
#> - Scenarios: ssp126 | ssp585
#> - GCMs: ACCESS-CM2 | MIROC6
#> All variables are located in the following root directory:
#> /tmp/Rtmpw1qyx4If we check the structure of the prepared_projection
object, we can see it’s a list containing:
- Paths to all variables representing distinct scenarios in subfolders.
- The pattern used to identify the format of raster files within the
folders (by default,
*.tif). - The names of the predictors.
- A list of class
prcompif a Principal Component Analysis (PCA) was performed on the set of variables withprepare_data().
#Open prepared_projection in a new window
View(pr)Project selected models to multiple scenarios
After preparing the data, we can use the
project_selected() function to predict the selected models
across the multiple scenarios specified in
prepare_projections:
## Create a folder to save projection results
#Here, in a temporary directory
out_dir <- file.path(tempdir(), "Projection_results/maxnet")
dir.create(out_dir, recursive = TRUE)
## Project selected models to multiple scenarios
p <- project_selected(models = fitted_model_maxnet,
projection_data = pr,
out_dir = out_dir,
write_partitions = TRUE,
progress_bar = FALSE) #Do not print progress barThe function returns a model_projections object. This
object is similar to the prepared_data object, storing
information on the predicted scenarios and the folder where the
resulting projection rasters were saved.
print(p)
#> model_projections object summary
#> ================================
#> Models projected for Present and Future
#> Future projections contain the following periods, scenarios and GCMs:
#> - Periods: 2041-2060 | 2081-2100
#> - Scenarios:
#> - GCMs: ACCESS-CM2 | MIROC6
#> All raster files containing the projection results are located in the following root directory:
#> /tmp/Rtmpw1qyx4/Projection_results/maxnetNote that the results were saved hierarchically in nested subfolders,
each representing a distinct scenario. In the root directory, the
function also saves a file named “Projection_paths.RDS”, which is the
model_projections object. This object can be imported into
R using
readRDS(file.path(out_dir, "Projection_paths.RDS")).
dir_tree(out_dir)
#> Temp\Projection_results/maxnet
#> ├── Future
#> │ ├── 2041-2060
#> │ │ ├── ssp126
#> │ │ │ ├── ACCESS-CM2
#> │ │ │ │ ├── General_consensus.tif
#> │ │ │ │ ├── Model_192_consensus.tif
#> │ │ │ │ ├── Model_192_partitions.tif
#> │ │ │ │ ├── Model_219_consensus.tif
#> │ │ │ │ └── Model_219_partitions.tif
#> │ │ │ └── MIROC6
#> │ │ │ ├── General_consensus.tif
#> │ │ │ ├── Model_192_consensus.tif
#> │ │ │ ├── Model_192_partitions.tif
#> │ │ │ ├── Model_219_consensus.tif
#> │ │ │ └── Model_219_partitions.tif
#> │ │ └── ssp585
#> │ │ ├── ACCESS-CM2
#> │ │ │ ├── General_consensus.tif
#> │ │ │ ├── Model_192_consensus.tif
#> │ │ │ ├── Model_192_partitions.tif
#> │ │ │ ├── Model_219_consensus.tif
#> │ │ │ └── Model_219_partitions.tif
#> │ │ └── MIROC6
#> │ │ ├── General_consensus.tif
#> │ │ ├── Model_192_consensus.tif
#> │ │ ├── Model_192_partitions.tif
#> │ │ ├── Model_219_consensus.tif
#> │ │ └── Model_219_partitions.tif
#> │ └── 2081-2100
#> │ ├── ssp126
#> │ │ ├── ACCESS-CM2
#> │ │ │ ├── General_consensus.tif
#> │ │ │ ├── Model_192_consensus.tif
#> │ │ │ ├── Model_192_partitions.tif
#> │ │ │ ├── Model_219_consensus.tif
#> │ │ │ └── Model_219_partitions.tif
#> │ │ └── MIROC6
#> │ │ ├── General_consensus.tif
#> │ │ ├── Model_192_consensus.tif
#> │ │ ├── Model_192_partitions.tif
#> │ │ ├── Model_219_consensus.tif
#> │ │ └── Model_219_partitions.tif
#> │ └── ssp585
#> │ ├── ACCESS-CM2
#> │ │ ├── General_consensus.tif
#> │ │ ├── Model_192_consensus.tif
#> │ │ ├── Model_192_partitions.tif
#> │ │ ├── Model_219_consensus.tif
#> │ │ └── Model_219_partitions.tif
#> │ └── MIROC6
#> │ ├── General_consensus.tif
#> │ ├── Model_192_consensus.tif
#> │ ├── Model_192_partitions.tif
#> │ ├── Model_219_consensus.tif
#> │ └── Model_219_partitions.tif
#> ├── Present
#> │ └── Present
#> │ ├── General_consensus.tif
#> │ ├── Model_192_consensus.tif
#> │ ├── Model_192_partitions.tif
#> │ ├── Model_219_consensus.tif
#> │ └── Model_219_partitions.tif
#> └── Projection_paths.RDSBy default, for each scenario, the function computes consensus metrics (mean, median, range, and standard deviation) for each model across its partitions (if more than one model was selected), as well as a general consensus across all models.
Note that each selected model can also have partitions. By default,
the function does not output these individual partitions unless
write_partitions = TRUE is set. It is important to write
the partitions if you intend to compute the variability across them
using projection_variability(). For more details, check the
vignette on Explore
Variability and Uncertainty in Projections.
The function accepts several other parameters that control the
predictions in predict_selected(), such as the consensus to
compute, the extrapolation type (free extrapolation (E),
extrapolation with clamping (EC), and no extrapolation
(NE)), variables to clamp, and the format of prediction
values (raw, cumulative,
logistic, or the default cloglog). For more
details, consult the vignette for Predict models to single scenario.
Import rasters resulting from projections
The model_projections object stores only the paths to
the resultant rasters. To import the results, we can use the
import_projections() function. By default, it imports all
consensus metrics (“median”, “range”, “mean”, and “stdev”) and all
scenarios (time periods, SSPs, and GCMs) available in the
model_projections object. Let’s import the mean for all
scenarios:
#Import mean of each projected scenario
p_mean <- import_projections(projection = p, consensus = "mean")
#Plot all scenarios
plot(p_mean, cex.main = 0.8)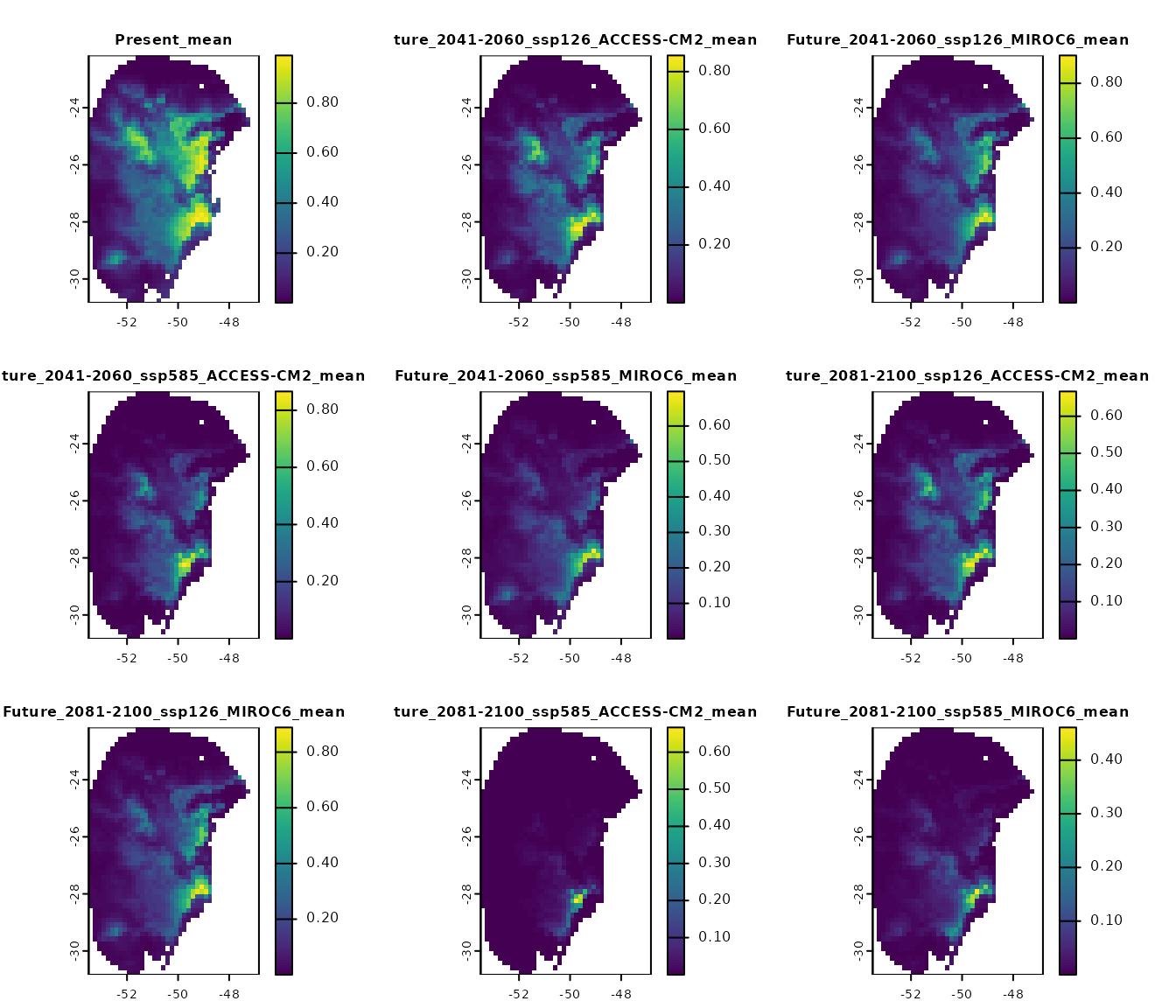
Alternatively, we can import results from specific scenarios. For example, let’s import the results only for the “2041-2060” time period under the SSP 126:
p_2060_ssp126 <- import_projections(projection = p, consensus = "mean",
present = FALSE, #Do not import present projections
future_period = "2041-2060",
future_pscen = "ssp126")
#Plot all scenarios
plot(p_2060_ssp126, cex.main = 0.8)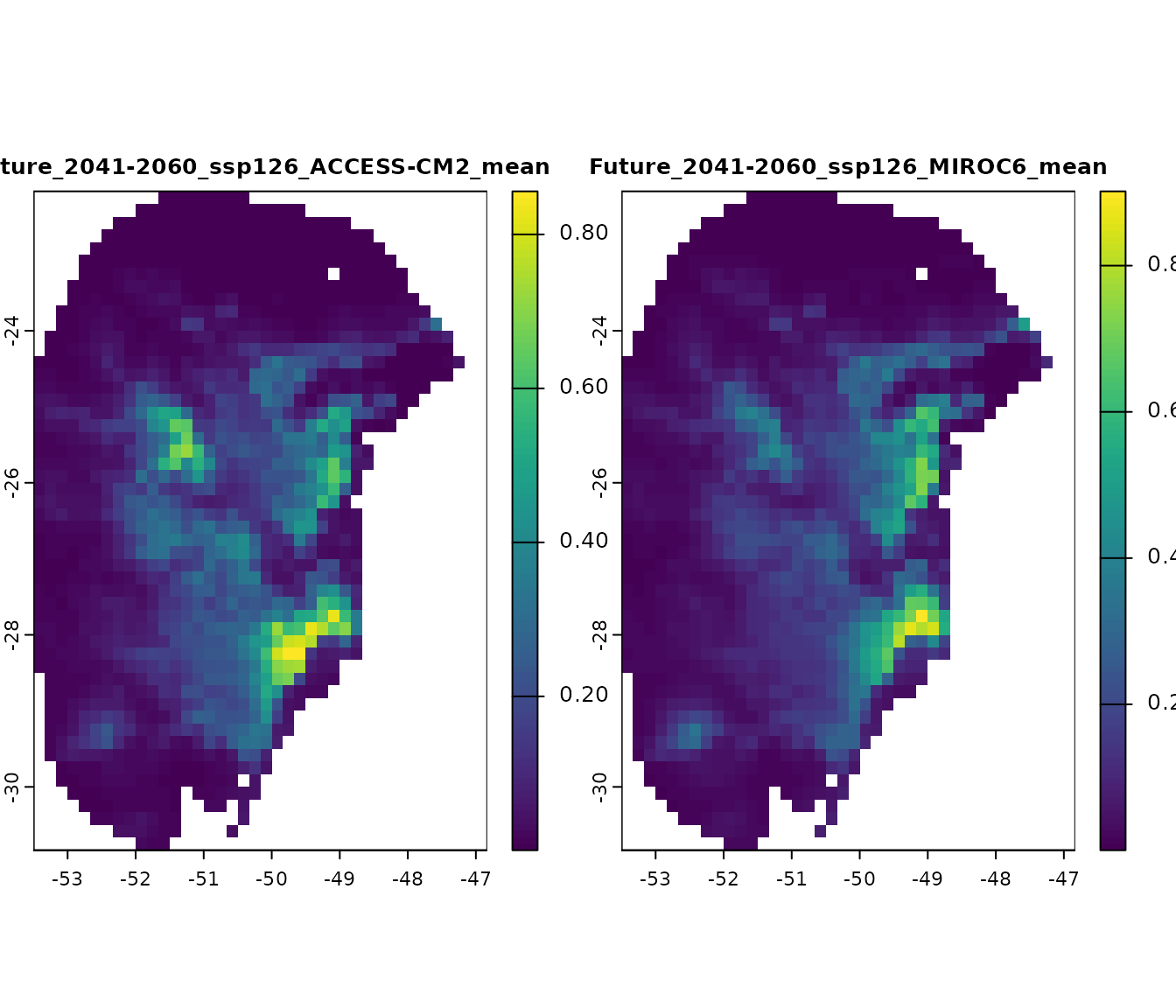
With the model_projections object, we can compute
changes in suitable areas between scenarios (see the
projection_changes function), explore variance stemming
from partitions, model parameterizations, and GCMs (see
projection_variability), and perform analysis of
extrapolation risks (see projection_mop). For more details,
check the vignette on Explore
Variability and Uncertainty in Projections.