Introduction
Model calibration is the most computationally challenging process
automated in kuenm2. In this step, candidate models are
trained and tested using a k-fold cross-validation approach. Then,
models are selected based on multiple criteria to warranty that the
models used in later steps are the most robust among the candidates. The
main function used in this step is calibration().
To start the calibration process, we need a
prepared_data object. For more details in data preparation,
please refer to the vignette prepare data
for model calibration.
To start, let’s create two prepared_data object: one
using the maxnet as algorithm, and another with GLM:
#Load packages
library(kuenm2)
library(terra)
#> terra 1.8.60
# Import occurrences
data(occ_data, package = "kuenm2")
# Import raster layers
var <- rast(system.file("extdata", "Current_variables.tif", package = "kuenm2"))
# Prepare data for maxnet model
d_maxnet <- prepare_data(algorithm = "maxnet",
occ = occ_data,
x = "x", y = "y",
raster_variables = var,
species = "Myrcia hatschbachii",
categorical_variables = "SoilType",
partition_method = "kfolds",
n_partitions = 4,
n_background = 1000,
features = c("l", "q", "lq", "lqp"),
r_multiplier = c(0.1, 1, 2))
# Prepare data for glm model
d_glm <- prepare_data(algorithm = "glm",
occ = occ_data,
x = "x", y = "y",
raster_variables = var,
species = "Myrcia hatschbachii",
categorical_variables = "SoilType",
partition_method = "bootstrap",
n_partitions = 10,
train_proportion = 0.7,
n_background = 300,
features = c("l", "q", "p", "lq", "lqp"),
r_multiplier = NULL) #Not necessary with glmsCalibration
The calibration() function fits and evaluates candidate
models considering the follow metrics:
- Omission error: calculated using models trained with separate testing data subsets. Users can specify multiple omission rates to be considered (e.g., c(5%, 10%)), though only one can be used as the threshold for selecting the best models.
- Partial ROC: calculated following Peterson et al. (2008).
- Model complexity (AIC): assessed using models generated with the complete set of occurrences.
- Unimodality (optional): Assessed through the beta coefficients of quadratic terms, following Arias-Giraldo & Cobos (2024).
In summary, to calibrate and evaluate the models, the function
requires a prepared_data object and the following
definitions:
- Omission Errors: Values ranging from 0 to 100, representing the percentage of potential error attributed to various sources of uncertainty in the data. These values are utilized in the calculation of omission rates and partial ROC.
- Omission Rate for Model Selection: The specific omission error threshold used to select models. This value defines the maximum omission rate a candidate model can have to be considered for selection.
- Removal of Concave Curves: A specification of whether to exclude candidate models that exhibit concave curves.
Optional arguments allow for modifications such as changing the delta
AIC threshold for model selection (default is 2), determining whether to
add presence samples to the background (default is TRUE),
and whether to employ user-specified weights. For a comprehensive
description of all arguments, refer to ?calibration.
In this example, we will evaluate the models considering two omission
errors (5% and 10%), with model selection based on the 5% omission
error. To improve computational speed, you can set parallel
to TRUE and specify the number of cores to utilize; the
candidate models will be distributed among these cores. To detect the
number of available cores on your machine, run
parallel::detectCores().
Maxnet Models
Let’s calibrate the maxnet models:
#Calibrate maxnet models
m_maxnet <- calibration(data = d_maxnet,
error_considered = c(5, 10),
omission_rate = 10,
parallel = FALSE, #Set TRUE to run in parallel
ncores = 1) #Define number of cores to run in parallel
# Task 1/1: fitting and evaluating models:
# |=====================================================================| 100%
#
# Model selection step:
# Selecting best among 300 models.
# Calculating pROC...
#
# Filtering 300 models.
# Removing 0 model(s) because they failed to fit.
# 135 models were selected with omission rate below 10%.
# Selecting 2 final model(s) with delta AIC <2.
# Validating pROC of selected models...
# |=====================================================================| 100%
# All selected models have significant pROC values.The calibration() function returns a
calibration_results object, a list containing various
essential pieces of information from the model calibration process. The
elements of the calibration_results object can be explored
by indexing them. For example, all evaluation metrics are stored within
the calibration_results element:
# See first rows of the summary of calibration results
head(m_maxnet$calibration_results$Summary[,c("ID", "Omission_rate_at_10.mean", "AICc",
"Is_concave")])
#> ID Omission_rate_at_10.mean AICc Is_concave
#> 1 1 0.0978 665.8779 FALSE
#> 2 2 0.0978 665.9493 FALSE
#> 3 3 0.0978 665.8956 FALSE
#> 4 4 0.1378 678.2084 FALSE
#> 5 5 0.1378 678.1407 FALSE
#> 6 6 0.1170 678.1182 FALSEWe can also examine the details of the selected models:
# See first rows of the summary of calibration results
m_maxnet$selected_models[,c("ID", "Formulas", "R_multiplier",
"Omission_rate_at_10.mean", "AICc", "Is_concave")]
#> ID
#> 192 192
#> 219 219
#> Formulas
#> 192 ~bio_1 + bio_7 + bio_15 + I(bio_1^2) + I(bio_7^2) + I(bio_15^2) -1
#> 219 ~bio_1 + bio_7 + bio_12 + bio_15 + I(bio_1^2) + I(bio_7^2) + I(bio_12^2) + I(bio_15^2) -1
#> R_multiplier Omission_rate_at_10.mean AICc Is_concave
#> 192 0.1 0.0769 608.8669 FALSE
#> 219 0.1 0.0962 610.0462 FALSEWhen printed, the calibration_results object provides a
summary of the model selection process. This includes the total number
of candidate models considered, the number of models that failed to fit,
and the number of models exhibiting concave curves (along with an
indication of whether these were removed). Additionally, it reports the
number of models excluded due to non-significant partial ROC (pROC)
values, high omission error rates, or elevated AIC values. Finally, a
summary of the metrics for the selected models is presented.
print(m_maxnet)
#> calibration_results object summary (maxnet)
#> =============================================================
#> Species: Myrcia hatschbachii
#> Number of candidate models: 300
#> - Models removed because they failed to fit: 0
#> - Models identified with concave curves: 39
#> - Model with concave curves not removed
#> - Models removed with non-significant values of pROC: 0
#> - Models removed with omission error > 10%: 165
#> - Models removed with delta AIC > 2: 133
#> Selected models: 2
#> - Up to 5 printed here:
#> ID
#> 192 192
#> 219 219
#> Formulas
#> 192 ~bio_1 + bio_7 + bio_15 + I(bio_1^2) + I(bio_7^2) + I(bio_15^2) -1
#> 219 ~bio_1 + bio_7 + bio_12 + bio_15 + I(bio_1^2) + I(bio_7^2) + I(bio_12^2) + I(bio_15^2) -1
#> Features R_multiplier pval_pROC_at_10.mean Omission_rate_at_10.mean
#> 192 lq 0.1 0 0.0769
#> 219 lq 0.1 0 0.0962
#> dAIC Parameters
#> 192 0.000000 6
#> 219 1.179293 7In this example, of the 300 candidate maxnet models fitted, two was selected based on a significant pROC value, a low omission error (<10%), and a low AIC score (<2).
GLM Models
Now, let’s calibrate the GLM Models to see if with this algorith we achieve different selected models:
#Calibrate maxnet models
m_glm <- calibration(data = d_glm,
error_considered = c(5, 10),
omission_rate = 10,
parallel = FALSE, #Set TRUE to run in parallel
ncores = 1) #Define number of cores to run in parallel
# Task 1/1: fitting and evaluating models:
# |=====================================================================| 100%
# Model selection step:
# Selecting best among 122 models.
# Calculating pROC...
#
# Filtering 122 models.
# Removing 0 model(s) because they failed to fit.
# 21 models were selected with omission rate below 10%.
# Selecting 1 final model(s) with delta AIC <2.
# Validating pROC of selected models...
# |=====================================================================| 100%
# All selected models have significant pROC values.Now, instead of two selected models, we have only one:
m_glm
#> calibration_results object summary (glm)
#> =============================================================
#> Species: Myrcia hatschbachii
#> Number of candidate models: 122
#> - Models removed because they failed to fit: 0
#> - Models identified with concave curves: 18
#> - Model with concave curves not removed
#> - Models removed with non-significant values of pROC: 0
#> - Models removed with omission error > 10%: 101
#> - Models removed with delta AIC > 2: 20
#> Selected models: 1
#> - Up to 5 printed here:
#> ID Formulas Features
#> 85 85 ~bio_1 + bio_7 + bio_12 + I(bio_1^2) + I(bio_7^2) + I(bio_12^2) lq
#> pval_pROC_at_10.mean Omission_rate_at_10.mean dAIC Parameters
#> 85 0 0.0904 0 6Concave curves
It is worth noting that with both maxnet and glm algorithm, some models were identified as having concave curves. Concave (or bimodal) curves indicate that the peak suitability is found at the extremes of the variable range. For example, as shown in the right panel of the figure below, higher suitability is observed in both the driest and wettest regions, with lower suitabilities occurring at intermediate precipitation levels.
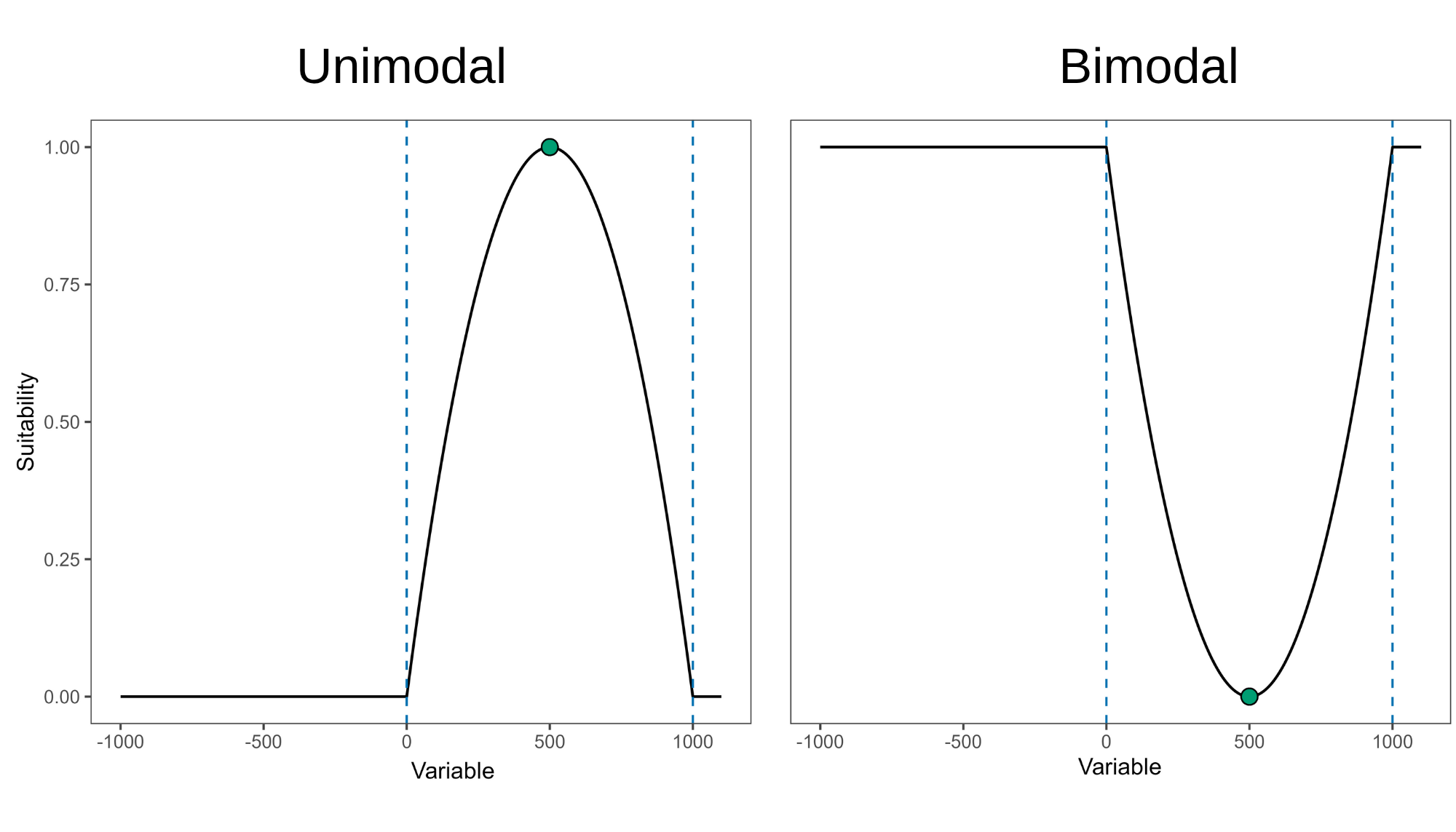
In our example, none of the selected models have concave curves:
#Selected maxnet models
m_maxnet$selected_models[,c("ID", "Formulas", "Is_concave")]
#> ID
#> 192 192
#> 219 219
#> Formulas
#> 192 ~bio_1 + bio_7 + bio_15 + I(bio_1^2) + I(bio_7^2) + I(bio_15^2) -1
#> 219 ~bio_1 + bio_7 + bio_12 + bio_15 + I(bio_1^2) + I(bio_7^2) + I(bio_12^2) + I(bio_15^2) -1
#> Is_concave
#> 192 FALSE
#> 219 FALSE
#Selected glm models
m_glm$selected_models[,c("ID", "Formulas", "Is_concave")]
#> ID Formulas
#> 85 85 ~bio_1 + bio_7 + bio_12 + I(bio_1^2) + I(bio_7^2) + I(bio_12^2)
#> Is_concave
#> 85 TRUEHowever, occasionally, a model with concave curves might be selected
if it has sufficiently low omission rate and AIC values. To
ensure that none of the selected models have concave
curves, you can set remove_concave = TRUE within
the calibration() function. Let’s test it with the maxnet
algorithm:
m_unimodal <- calibration(data = d_maxnet,
remove_concave = TRUE, # Ensures concave models are not selected
error_considered = c(5, 10),
omission_rate = 10)
# Task 1/2: checking for concave responses in models:
# |=====================================================================| 100%
#
# Task 2/2: fitting and evaluating models with no concave responses:
# |=====================================================================| 100%
#
# Model selection step:
# Selecting best among 300 models.
# Calculating pROC...
#
# Filtering 300 models.
# Removing 0 model(s) because they failed to fit.
# Removing 39 model(s) with concave curves.
# 110 models were selected with omission rate below 10%.
# Selecting 2 final model(s) with delta AIC <2.
# Validating pROC of selected models...
# |=====================================================================| 100%
# All selected models have significant pROC values.Note that the process is now divided into two tasks:
Task 1/2: Only candidate models that include quadratic terms are fitted. For Maxent models (using the maxnet algorithm), the function first fits the candidate model with the highest regularization multiplier (e.g., 5) for each formula. This approach is used because if a particular formula produces a concave response at a high regularization value, it will also produce concave responses at lower regularization values. By checking the model with the highest regularization first, the function can skip fitting the other models with the same formula but lower regularization values, saving time and computation.
-
Task 2/2: In this step, the function fits and evaluates two groups of models:
- Models without quadratic terms.
- Models with quadratic terms, but only those with formulas that did not produce concave responses in Task 1/2 (i.e., they passed the concavity check at the higher regularization multiplier).
Re-selecting models
The model selection procedure is conducted internally during the calibration process. However, it is possible to re-select models by considering other omission rates (since these were calculated during calibration), model complexity (delta AIC) and concave curves.
By default, calibration() calculates pROC values only
for the selected models to optimize computational time. Consequently,
pROC values for non-selected models are filled with NA.
# See first rows of the summary of calibration results (pROC values)
head(m_maxnet$calibration_results$Summary[,c("ID", "Mean_AUC_ratio_at_10.mean",
"pval_pROC_at_10.mean")])
#> ID Mean_AUC_ratio_at_10.mean pval_pROC_at_10.mean
#> 1 1 NA NA
#> 2 2 NA NA
#> 3 3 NA NA
#> 4 4 NA NA
#> 5 5 NA NA
#> 6 6 NA NA
# See pROC values of selected models
m_maxnet$selected_models[,c("ID", "Mean_AUC_ratio_at_10.mean",
"pval_pROC_at_10.mean")]
#> ID Mean_AUC_ratio_at_10.mean pval_pROC_at_10.mean
#> 192 192 1.497376 0
#> 219 219 1.502309 0When pROC is not calculated for all models during
calibration(), the select_models() function
requires the prepared_data used during the calibration
step, and compute_proc must be set to
TRUE.
For instance, let’s re-select the maxnet models from the calibration results, applying an omission rate of 5% instead 10%:
#Re-select maxnet models
new_m_maxnet <- select_models(calibration_results = m_maxnet,
data = d_maxnet, #Necessary for computing pROC
compute_proc = TRUE,
omission_rate = 5) # New omission rate
#> Selecting best among 300 models.
#> Calculating pROC...
#>
#> Filtering 300 models.
#> Removing 0 model(s) because they failed to fit.
#> 116 models were selected with omission rate below 5%.
#> Selecting 2 final model(s) with delta AIC <2.
#> Validating pROC of selected models...
#>
#> All selected models have significant pROC values.
print(new_m_maxnet)
#> calibration_results object summary (maxnet)
#> =============================================================
#> Species: Myrcia hatschbachii
#> Number of candidate models: 300
#> - Models removed because they failed to fit: 0
#> - Models identified with concave curves: 39
#> - Model with concave curves not removed
#> - Models removed with non-significant values of pROC: 0
#> - Models removed with omission error > 5%: 184
#> - Models removed with delta AIC > 2: 114
#> Selected models: 2
#> - Up to 5 printed here:
#> ID Formulas
#> 159 159 ~bio_1 + bio_7 + I(bio_1^2) + I(bio_7^2) -1
#> 189 189 ~bio_1 + bio_7 + bio_12 + I(bio_1^2) + I(bio_7^2) + I(bio_12^2) -1
#> Features R_multiplier pval_pROC_at_5.mean Omission_rate_at_5.mean dAIC
#> 159 lq 0.1 0 0.0192 0.8581936
#> 189 lq 0.1 0 0.0192 0.0000000
#> Parameters
#> 159 4
#> 189 6If a calibration_results object is provided,
select_models() will return a
calibration_results output with the selected models and
summary updated. Note that we now have different selected models with
the maxnet algoritm:
new_m_maxnet$selected_models[,c("ID", "Formulas", "R_multiplier",
"Omission_rate_at_5.mean", "Mean_AUC_ratio_at_5.mean",
"AICc", "Is_concave")]
#> ID Formulas
#> 159 159 ~bio_1 + bio_7 + I(bio_1^2) + I(bio_7^2) -1
#> 189 189 ~bio_1 + bio_7 + bio_12 + I(bio_1^2) + I(bio_7^2) + I(bio_12^2) -1
#> R_multiplier Omission_rate_at_5.mean Mean_AUC_ratio_at_5.mean AICc
#> 159 0.1 0.0192 1.480830 622.7677
#> 189 0.1 0.0192 1.513242 621.9095
#> Is_concave
#> 159 FALSE
#> 189 FALSEYou can also provide a data.frame containing the
evaluation metrics for each candidate model directly to
select_models(). This data.frame is available
in the output of the calibration() function under
object$calibration_results$Summary. In this case, the
function will return a list containing the selected models along with
summaries of the model selection process.
#Re-select models using data.frame
new_summary <- select_models(candidate_models = m_maxnet$calibration_results$Summary,
data = d_maxnet, #Necessary for computing pROC
compute_proc = TRUE,
omission_rate = 5)
#> Selecting best among 300 models.
#> Calculating pROC...
#>
#> Filtering 300 models.
#> Removing 0 model(s) because they failed to fit.
#> 116 models were selected with omission rate below 5%.
#> Selecting 2 final model(s) with delta AIC <2.
#> Validating pROC of selected models...
#>
#> All selected models have significant pROC values.
#Get class of object
class(new_summary)
#> [1] "list"
#See selected models
new_summary$selected_models[,c("ID", "Formulas", "R_multiplier",
"Omission_rate_at_5.mean", "Mean_AUC_ratio_at_5.mean",
"AICc", "Is_concave")]
#> ID Formulas
#> 159 159 ~bio_1 + bio_7 + I(bio_1^2) + I(bio_7^2) -1
#> 189 189 ~bio_1 + bio_7 + bio_12 + I(bio_1^2) + I(bio_7^2) + I(bio_12^2) -1
#> R_multiplier Omission_rate_at_5.mean Mean_AUC_ratio_at_5.mean AICc
#> 159 0.1 0.0192 1.479831 622.7677
#> 189 0.1 0.0192 1.514019 621.9095
#> Is_concave
#> 159 FALSE
#> 189 FALSESaving a calibration_results object
After calibrating and selecting the best-performing models, we can
proceed to fit the final models (see the vignette for model exploration)
using the calibration_results object. As this object is
essentially a list, users can save it to a local directory using
saveRDS(). Saving the object facilitates loading it back
into your R session later using readRDS(). See an example
below: